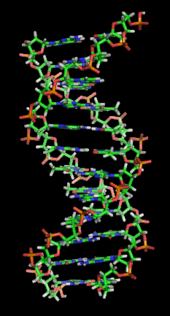
DNA is a long polymer made from repeating units called nucleotides. As first discovered by James D. Watson and Francis Crick, the structure of DNA of all species comprises two helical chains each coiled round the same axis, and each with a pitch of 34 Ångströms (3.4 nanometres) and a radius of 10 Ångströms(1.0 nanometres).According to another study, when measured in a particular solution, the DNA chain measured 22 to 26 Ångströms wide (2.2 to 2.6 nanometres), and one nucleotide unit measured 3.3 Å (0.33 nm) long.Although each individual repeating unit is very small, DNA polymers can be very large molecules containing millions of nucleotides. For instance, the largest human chromosome, chromosome number 1, is approximately 220 million base pairslong.
In living organisms DNA does not usually exist as a single molecule, but instead as a pair of molecules that are held tightly together. These two long strands entwine like vines, in the shape of a double helix. The nucleotide repeats contain both the segment of the backbone of the molecule, which holds the chain together, and a nucleobase, which interacts with the other DNA strand in the helix. A nucleobase linked to a sugar is called a nucleoside and a base linked to a sugar and one or more phosphate groups is called a nucleotide. Polymers comprising multiple linked nucleotides (as in DNA) is called a polynucleotide.
The backbone of the DNA strand is made from alternating phosphate and sugarresidues. The sugar in DNA is 2-deoxyribose, which is a pentose (five-carbon) sugar. The sugars are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These asymmetric bonds mean a strand of DNA has a direction. In a double helix the direction of the nucleotides in one strand is opposite to their direction in the other strand: the strands are antiparallel. The asymmetric ends of DNA strands are called the 5′ (five prime) and 3′ (three prime) ends, with the 5' end having a terminal phosphate group and the 3' end a terminal hydroxyl group. One major difference between DNA and RNA is the sugar, with the 2-deoxyribose in DNA being replaced by the alternative pentose sugar ribose in RNA.

A section of DNA. The bases lie horizontally between the two spiraling strands.Animated version at File:DNA orbit animated.gif.
The DNA double helix is stabilized primarily by two forces: hydrogen bonds between nucleotides and base-stacking interactions among the aromatic nucleobases. In the aqueous environment of the cell, the conjugated π bonds of nucleotide bases align perpendicular to the axis of the DNA molecule, minimizing their interaction with the solvation shell and therefore, the Gibbs free energy. The four bases found in DNA are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). These four bases are attached to the sugar/phosphate to form the complete nucleotide, as shown for adenosine monophosphate.
The nucleobases are classified into two types: the purines, A and G, being fused five- and six-memberedheterocyclic compounds, and the pyrimidines, the six-membered rings C and T. A fifth pyrimidine nucleobase, uracil (U), usually takes the place of thymine in RNA and differs from thymine by lacking amethyl group on its ring. Uracil is not usually found in DNA, occurring only as a breakdown product of cytosine. In addition to RNA and DNA a large number of artificial nucleic acid analogues have also been created to study the proprieties of nucleic acids, or for use in biotechnology.