Nucleic acids have a primary, secondary, and tertiary structure analogous to the classification of protein structure. The sequence of bases in the nucleic acid chain gives the primary structure of DNA or RNA. The sequence of bases is read in a 5′ → 3′ direction, so that you would read the structure in the next figure as ACGT. See Figure
1 .
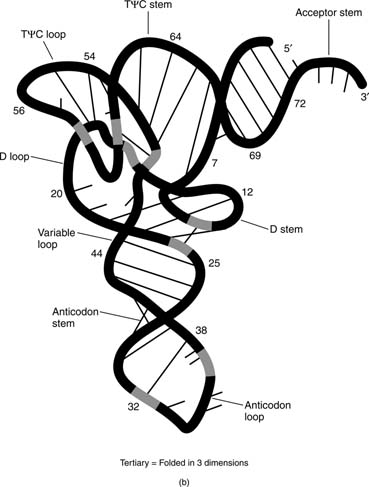
The regions of the secondary structure do not have to form between sequences that are close together. For example, base-pairing between the sequences at the 5′ and 3′ ends forms the acceptor stem of transfer RNA. The tertiary structure of a nucleic acid refers to the three-dimensional arrangement of the nucleic acid—that is, the arrangement of the molecule in space, as in the tertiary structure of tRNA.
Sugar structures in DNA and RNA
Although RNA and DNA are similar in their overall properties, the presence of the 2′-hydroxyl group of ribose in RNA and its replacement by 2′-deoxyribose in DNA make a large contribution to the different chemical properties of DNA and RNA. The 2′-hydroxyl group makes RNA susceptible to alkaline hydrolysis in solutions of pH greater than nine or so. Conversely, the bonds between the sugar and the purine bases of DNA are more easily broken in low pH conditions than are those of RNA.
The 2′-hydroxyl group affects the tertiary structure of RNA. First, the conformation of the sugar is different between DNA and RNA. Secondly, the 2′-hydroxyl group provides hydrogen bond donor and acceptor functions for formation of hydrogen bonds. These hydrogen bonds are important in the formation of the tertiary structure of an RNA and are not available to DNA. Although single-stranded DNA does have some tertiary structure, this structure is usually not as stable as that of an RNA of the same sequence.
DNA structure
The Watson-Crick base-pairing of the two strands largely determines thesecondary structure of DNA. All naturally occurring DNAs are double-stranded, for at least some of their lifetimes. Double-stranded DNA is a fairly uniform structure, and the need for a regular structure is one way in which changes in DNA (genetic mutations) can be detected. The fact that A-T base pairs and G-C base pairs have very similar sizes means that no “bulges” or “gaps” exist within the double helix. An irregular place in the double helix means that something is wrong with the structure, and this signals the need for DNA repair systems to fix the damage.
The A-T base pair has two hydrogen bonds; each base serves as H-donor for one bond and as H-acceptor for the other.
Melting and helix formation of nucleic acids are often detected by the absorbance of ultraviolet light. This process can be understood in the following way: The stacked bases shield each other from light. As a result, the absorbance of UV light whose wavelength is 260 nanometers (the A260) of a double-helical DNA is less than that of the same DNA, whose strands are separated (the random coil). This effect is called the hypochromicity (less-color) of the double-helical DNA.
If a double-stranded DNA is heated, the strands separate. The temperature at which the DNA is halfway between the double-stranded and the random structure is called the
melting temperature (Tm) of that DNA. The T
m of a DNA depends on base composition. G-C base pairs are stronger than A-T base pairs; therefore, DNAs with a high G+C content have a higher T
m than do DNAs with a higher A+T content. For example, human DNA, which is close to 50 percent G+C, might melt at 70°, while DNA from the bacterium
Streptomyces, which has close to 73 percent G+C, might melt at 85°. The T
m of a DNA also depends on solvent composition. High ionic strength—for example, a high concentration of NaCl—promotes the double-stranded state (raises the T
m) of a given DNA because the higher concentration of positive sodium ions masks the negative charge of the phosphates in the DNA backbone. Finally, the T
m of a DNA depends on how well its bases match up. A synthetic DNA double strand made with some mismatched base-pairs has a lower T
m compared to a completely double-stranded DNA. This last property is important in using DNA from one species to detect similar DNA sequences of another species. For example, the DNA coding for an enzyme from human cells can form double helices with mouse DNA sequences coding for the same enzyme; however, the mouse-mouse and human-human double strands will both melt at a higher temperature than will the human-mouse hybrid DNA double helices.
Direct reactions with DNA serve as the molecular basis for the action of several anti-tumor drugs. Cancer is primarily a disease of uncontrolled cell growth, and cell growth depends on DNA synthesis. Cancer cells are often more sensitive than normal cells to compounds that damage DNA. For example, the anti-tumor drug cisplatin reacts with guanine bases in DNA and the daunomycin antibiotics act by inserting into the DNA chain between base pairs. In either case, these biochemical events can lead to the death of a tumor cell.
DNA tertiary structure
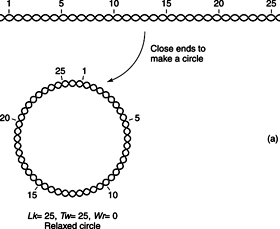
The DNA double helix may be arranged in space, in a tertiary arrangement of the strands. The two strands of DNA wind around each other. In a covalently closed circular DNA, this means that the two strands can't be separated. Because the DNA strands can't be separated, the total number of turns in a given molecule of closed circular DNA is a constant, called the Linking Number, or Lk. The linking number of a DNA is an integer and has two components, the Twist ( Tw), or number of helical turns of the DNA, and the Writhe ( Wr), or the number ofsupercoiled turnsin the DNA. Because L is a constant, the relationship can be shown by the equation:
Figures 5a and
5b , which show a double helical DNA with a linking number equal to 23, best illustrate this equation.
Normally, this DNA would have a linking number equal to 25, so it is
underwound. The DNA double helical structures in the previous figure have the same value of Lk; however, the DNA can be supercoiled, with the two “underwindings” taken up by the negative supercoils. This is equivalent to two “turns'-worth” of single-stranded DNA and no supercoils. This interconversion of helical and superhelical turns is important in gene transcription and regulation.
Enzymes called DNA topoisomerases alter Lk, the linking number of a DNA, by a bond breaking and rejoining process. Naturally-occurring DNAs have negative supercoils; that is, they are “underwound.” Type I topoisomerases (sometimes called “nicking-closing enzymes”) carry out the conversion of negatively supercoiled DNA to relaxed DNA in increments of one turn. That is, they increase Lk by increments of one to a final value of zero. Type I topoisomerases are energy independent, because they don't require ATP for their reactions. Some anti-tumor drugs, including campothecin, target the eukaryotic topoisomerase I enzyme. Type II topoisomerases (sometimes called DNA gyrases) reduce Lk by increments of two. These enzymes are ATP-dependent and will alter the linking number of any closed circular DNA. The antibiotic naladixic acid, which is used to treat urinary tract infections, targets the prokaryotic enzyme. Type II topoisomerases act on naturally occurring DNAs to make them supercoiled. Topoisomerases play an essential role in DNA replication and transcription.